Protein Sequences and Antibotics¶
- Alexander Fleming discovered Antibotics in 1928
- Small peptide sequences that target and kill bacteria
Generated naturally by fungi and other bacteria
Today's questions
- Where do they come from?
- How can one infer their sequence?
1
Discovery of Penicillin (1928)¶

- Upon returning from holiday in 1928 a british biologist and pharmacolgist, Alexander Fleming, discovered a strange pattern of cell death in a stack of staphylococci cultures he had on a bench in a corner of his laboratory.
- Fleming noticed that one culture was contaminated with a fungus, and that the colonies of staphylococci immediately surrounding the fungus had been destroyed, whereas other staphylococci colonies farther away were normal, and famously remarked "That's funny".
- He attempted to isolate the agent that killed the bacteria, and even hypothesized that such an agent might be suitable for treating infections known to be caused by staphylococci. But, he had little luck.
- It proved hard to collect enough Penicillin to make it viable af a drug throughout the 1930s.
- Eventually, a mouldy cantalope found in Illinois in 1943, would provide a mold capable of generating high quantities of Penicillin and greatly reduce the World War II battlefield infection called Sepsis.
- The chemical structure of Penicillin was not determined until 1945 by Dorothy Hodgkin, another british X-ray crystallographer like Rosalind Franklin.
2
Other Antibotics¶

- Meanwhile, a second important antibotic, Gramicidin S or Gramicidin Soviet, was discovered by Russian microbiologist Georgyi Gause and his wife Maria Brazhnikova in 1942. This antibotic was discovered in another strain of bacteria, Bacillus brevis, which had the tendancy to kill staphylococci nearby it.
- It was actually easier to mass-produce than Penicillin, and it saved the lives of many russians in the second world war, as well as the lives of its inventors.
- Gramicidin S is largly composed of a cyclodecapeptide, a protein-like peptide chain composed of 10 amino-acids joined head-to-tail to form a ring, called Tyrocidine B1.
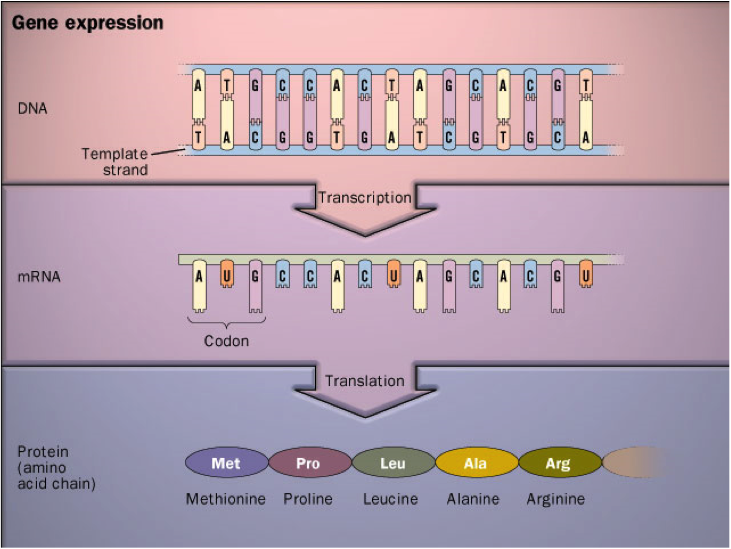
- The Central Dogma of Molecular Biology suggests that templates for such peptide chains are encoded in DNA.
- Our first task today is to see if we can find the genomic region where Gramicidin S is encoded.
3
Recall the Central Dogma from Lecture 1¶
Francis Crick, one of the discoverers of DNA structure, originally coined the term "Central Dogma" in 1958. That choice of words was particularly provocative.
 Francis Crick |
 |
4
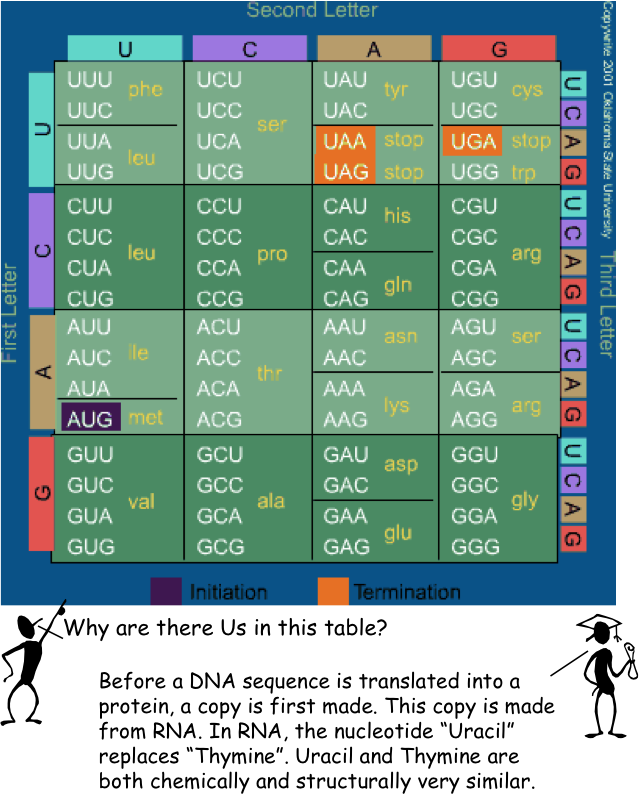
DNA Transcription¶
|
|
5
Tyrocidine B1's Peptide Sequence¶
Tyrocidine B1 is a small peptide composed of only 10 amino acids.
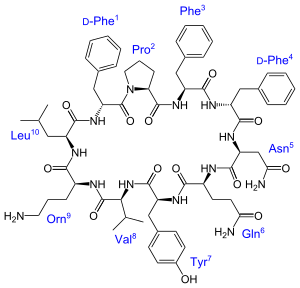
Amino Acid: Valine Leucine Proline Phenylalanine Glutamine 3-letter Abbr: Val-Lys-Leu-Phe-Pro-Trp-Phe-Asn-Gln-Tyr
1-letter Abbr: V K L F P W F N Q Y Amino Acid: Lysine Phenylalanine Tryptophan Asparagine Tyrosine
Also note that the Lysine is modified during the circularization into another non-proteinogenic amino acid, Ornithine.
6
A toolkit of Python Dictionarys¶
- The following Python Dictionaries will be used to aid our search
codon = {
"AAA": 'K', "AAG": 'K', "AAC": 'N', "AAU": 'N',
"AGA": 'R', "AGG": 'R', "AGC": 'S', "AGU": 'S',
"ACA": 'T', "ACG": 'T', "ACC": 'T', "ACU": 'T',
"AUA": 'I', "AUG": 'M', "AUC": 'I', "AUU": 'I',
"GAA": 'E', "GAG": 'E', "GAC": 'D', "GAU": 'D',
"GGA": 'G', "GGG": 'G', "GGC": 'G', "GGU": 'G',
"GCA": 'A', "GCG": 'A', "GCC": 'A', "GCU": 'A',
"GUA": 'V', "GUG": 'V', "GUC": 'V', "GUU": 'V',
"CAA": 'Q', "CAG": 'Q', "CAC": 'H', "CAU": 'H',
"CGA": 'R', "CGG": 'R', "CGC": 'R', "CGU": 'R',
"CCA": 'P', "CCG": 'P', "CCC": 'P', "CCU": 'P',
"CUA": 'L', "CUG": 'L', "CUC": 'L', "CUU": 'L',
"UAA": '*', "UAG": '*', "UAC": 'Y', "UAU": 'Y',
"UGA": '*', "UGG": 'W', "UGC": 'C', "UGU": 'C',
"UCA": 'S', "UCG": 'S', "UCC": 'S', "UCU": 'S',
"UUA": 'L', "UUG": 'L', "UUC": 'F', "UUU": 'F'
}
AminoAcid = {
'A': 'Alanine', 'C': 'Cysteine', 'D': 'Aspartic acid', 'E': 'Glutamic acid',
'F': 'Phenylalanine', 'G': 'Glycine', 'H': 'Histidine', 'I': 'Isoleucine',
'K': 'Lysine', 'L': 'Leucine', 'M': 'Methionine', 'N': 'Asparagine',
'P': 'Proline', 'Q': 'Glutamine', 'R': 'Arginine', 'S': 'Serine',
'T': 'Theronine', 'V': 'Valine', 'W': 'Tryptophan', 'Y': 'Tyrosine',
'*': 'STOP'
}
AminoAbbrv = {
'A': 'Ala', 'C': 'Cys', 'D': 'Asp', 'E': 'Glu',
'F': 'Phe', 'G': 'Gly', 'H': 'His', 'I': 'Ile',
'K': 'Lys', 'L': 'Leu', 'M': 'Met', 'N': 'Asn',
'P': 'Pro', 'Q': 'Gln', 'R': 'Arg', 'S': 'Ser',
'T': 'Thr', 'V': 'Val', 'W': 'Trp', 'Y': 'Tyr',
'*': 'STP'
}
# We'll use this dictionary later
Daltons = {
'A': 71, 'C': 103, 'D': 115, 'E': 129,
'F': 147, 'G': 57, 'H': 137, 'I': 113,
'K': 128, 'L': 113, 'M': 131, 'N': 114,
'P': 97, 'Q': 128, 'R': 156, 'S': 87,
'T': 101, 'V': 99, 'W': 186, 'Y': 163
}
7
Now our Peptide sequence¶
We'll use the 1-letter Amino Acid abbreviations to represent our sequence.
TyrocidineB1 = "VKLFPWFNQY"
Now, let's derive a new dictionary that provides for every Amino acid the set of codons that can encode it.
TrimerCodes = {}
for key, code in codon.iteritems():
TrimerCodes[code] = TrimerCodes.get(code,[]) + [key]
for key in sorted(TrimerCodes.iterkeys()):
print "%15s (%1s): %s" % (AminoAcid[key], key, TrimerCodes[key])
8
How many ways to code Tyrocidine B1?¶
- Since most Amino Acids have multiple codon encodings, any given sequence of Amino Acids can be codes in mltiple ways.
- Let's figure out exactly how many ways there are to encode our little peptide.
codes = 1
for residue in TyrocidineB1:
print residue, AminoAbbrv[residue], TrimerCodes[residue]
codes *= len(TrimerCodes[residue])
print "%d possible sequences" % codes
9
Where do we start?¶
- Consider that translation could begin at any point in the genome.
- Genes can be encoded on either strand
- There are 3 possible open-read frames (mod-3 starting positions) on each strand.
- Meanwhile, our peptide has 10 possible starting positions, since it is circular we can't be sure which codon appears first.
Let's start with something simpler¶
- Converting a DNA sequence into a protein sequence.
- In other words, the computational version of translation
def revComp(dnaSeq):
return ''.join([{'A':'T','C':'G','G':'C','T':'A'}[base] for base in reversed(dnaSeq)])
def proteinTranslation(dnaSeq):
rnaSeq = dnaSeq.replace("T", "U")
protSeq = ''
for i in xrange(0,len(dnaSeq),3):
if (i+3 > len(dnaSeq)):
break
protSeq += AminoAbbrv[codon[rnaSeq[i:i+3]]]
return rnaSeq, protSeq
10
Let's test it¶
# one way to code TyrocidineB1
DNA = 'GTGAAACTTTTTCCTTGGTTTAATCAATAT'
DNAr = revComp(DNA)
print "A short DNA sequence"
print "5'-%s-3'" % DNA
print "3'-%s-5'" % ''.join([{'A':'T','C':'G','G':'C','T':'A'}[base] for base in DNA])
print
print "Read frames in primary sequence"
for frame in xrange(3):
RNA, Peptides = proteinTranslation(DNA[frame:])
print "%d %s" % (frame+1,DNA)
print "%s %s" % (frame*" ", RNA)
print "%s %s" % (frame*" ", Peptides)
print
print
print "Read frames in the reverse-complement sequence"
for frame in xrange(3):
RNA, Peptides = proteinTranslation(DNAr[frame:])
print "%d %s" % (frame+1,DNAr)
print "%s %s" % (frame*" ", RNA)
print "%s %s" % (frame*" ", Peptides)
print
11
Now let's try Something Real¶
def loadFasta(filename):
""" Parses a classically formatted and possibly
compressed FASTA file into a list of headers
and fragment sequences for each sequence contained"""
if (filename.endswith(".gz")):
fp = gzip.open(filename, 'rb')
else:
fp = open(filename, 'rb')
# split at headers
data = fp.read().split('>')
fp.close()
# ignore whatever appears before the 1st header
data.pop(0)
headers = []
sequences = []
for sequence in data:
lines = sequence.split('\n')
headers.append(lines.pop(0))
# add an extra "+" to make string "1-referenced"
sequences.append('+' + ''.join(lines))
return (headers, sequences)
header, seq = loadFasta("data/BacillusBrevis.fa")
for i in xrange(len(header)):
print header[i]
print len(seq[i])-1, "bases", seq[i][:30], "...", seq[i][-30:]
print
Over 6 million bases to look through!
12
Some simple "Computational" experiments¶
- Just like a biologist, it is ill-advised to jump into a problem without first getting some sense of what works and what doesn't. Let's first take a look at our data
- One thing you might have noticed about the possible encodings of Tyrocidine B1 is that there is a single Tryptophan and only one encoding for this amino acid
- Let's try to anchor our search around that seed and grow from there.
Step 1: Where and how many Tryptophan encodings¶
def CodonSeqCount(genome, codonSeq):
N = 0
start = 0
while True:
pos = genome.find(codonSeq, start)
if (pos < 0):
break
else:
N += 1
start = pos + 1
return N
genome = seq[0]
tryptophanCode = TrimerCodes['W'][0].replace('U','T')
print "Searching for", tryptophanCode,
print "Found it", CodonSeqCount(genome,tryptophanCode), "times"
revCompCode = revComp(tryptophanCode)
print "Searching for", revCompCode,
print "Found it", CodonSeqCount(genome,revCompCode), "times"
Is this about what you expected? $\frac {6.3 \times 10^6}{64} \approx 100,000$. We've narrowed our search some, but there's still alot.
13
Narrowing things down more¶
- Rather than searching for every possible string, let's examine the codons around this initial anchor.
def AminoAcidSeqCount(genome, peptideSeq):
# Note: Only checks primary strand
readframe = []
anchor = peptideSeq[0]
for rnaSeq in TrimerCodes[anchor]:
dnaSeq = rnaSeq.replace('U','T')
start = 0
while True:
pos = genome.find(dnaSeq, start)
if (pos < 0):
break
else:
i = 1
while (i < len(peptideSeq) and (pos+3*(i+1) <= len(genome))):
if (codon[genome[pos+3*i:pos+3*(i+1)].replace('T','U')] != peptideSeq[i]):
break
i += 1
else:
readframe.append(pos)
start = pos + 1
return readframe
peptide = "WFNQY"
for i in xrange(1,len(peptide)+1):
ORFs = AminoAcidSeqCount(genome,peptide[:i])
print "Searching for", peptide[:i],
print "Found it", len(ORFs), "times"
14
Could we have been Lucky this time?¶
Let's take a look:
for pos in ORFs:
start = pos
end = pos + (3*len(peptide))
print "%9d %s %s" % (start, genome[start:end], proteinTranslation(genome[start:end])[1])
Recall we are looking for a substring of ValLysLeuPheProTrpPheAsnGlnTyr, which we found, but to be sure let's look around a little more.
for pos in ORFs:
start = pos - (2*3)
end = pos + (3*(len(peptide)+2))
print "%9d %s %s" % (start, genome[start:end], proteinTranslation(genome[start:end])[1])
We didn't find what we were expecting.
15
What Went Wrong?¶
- We only searched in one direction from our anchor
- We didn't search the reverse-complement sequence
- We didn't consider that the cycle could have been broken somewhere in between "W-F-N-Q-Y"
- One of the earlier 5, 111, 3922, or 106555 candidates might be the solution
- The best approach might be to call AminoAcidSeqCount() will all 10 circular permutations of "VKLFPWFNQY" (assuming that we also fix the reverse-complement sequence problem)
[TyrocidineB1[i:]+TyrocidineB1[:i] for i in xrange(len(TyrocidineB1))]
But even if you tried alll these fixes, you would still not find the code for Tyrocidine B1 in Bacillus brevis.
16
Proteins that make Petide Chains¶
Bacteria and fungi use large multifunctional enzymes called NonRibosomal Peptide Synthetases (NRPSs) to produce peptide chains that are not encoded in DNA. Nonribosomal peptides are synthesized by nonribosomal peptide synthetases, which, unlike the ribosomes, are independent of messenger RNA. Each nonribosomal peptide synthetase can synthesize only one type of peptide. They are often toxins, siderophores, or pigments. Nonribosomal peptide antibiotics, cytostatics, and immunosuppressants are widely used as drugs.
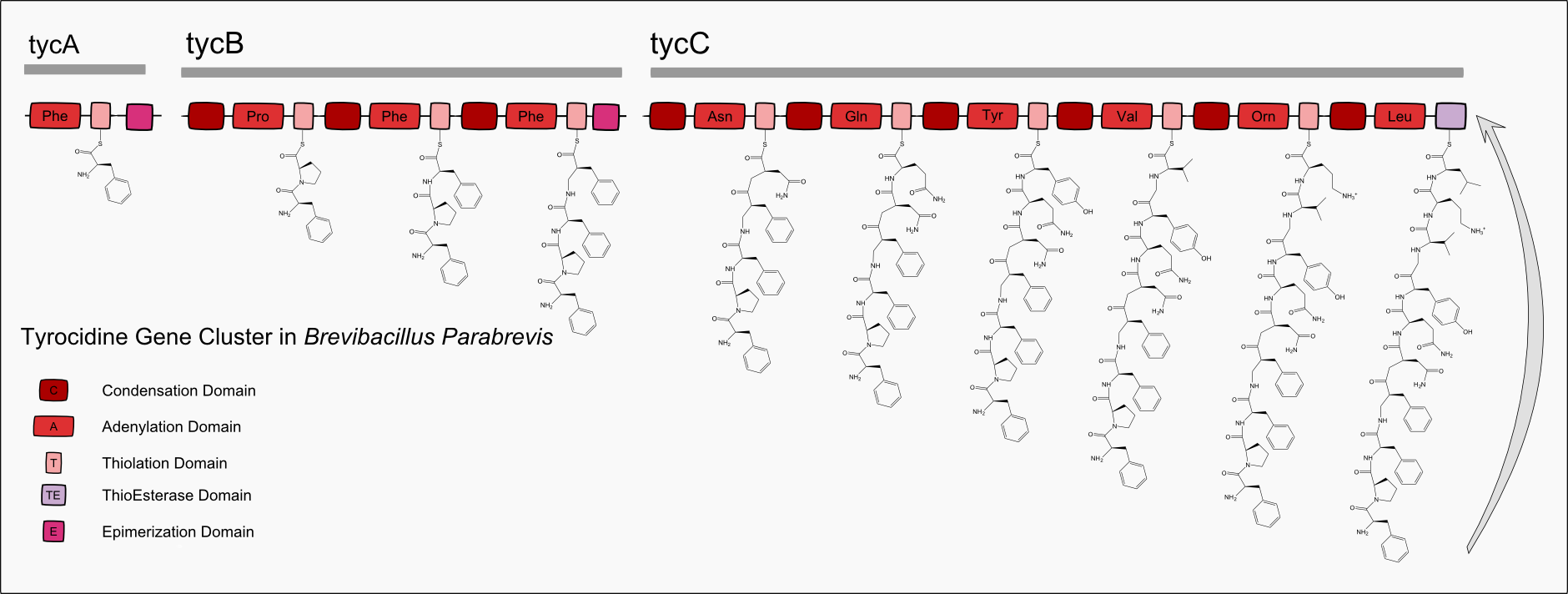
17
So we are left with a Question¶
- How do you figure out a peptide's amino acid sequence? It may not appear in the genome.
- Next time we will delve deeper into laboratory methods to start a discussion about peptide sequence inference

18