Finding Hidden Patterns in DNA¶
- What makes searching for frequent subsequences hard?
- Allowing for errors?
- All the places they could be hiding?
- Will Thursday Night work for the Python Tutorial?
1
Initiating Transcription¶
- As a precursor to transcription (the reading of DNA to construct RNAs, eventually leading to protein synthesis) special proteins bind to the DNA, and separate it to enable its reading.
- How do these proteins know where the coding genes are in order to bind?

- Genes are relatively rare
- O(1,000,000,000) bases/genome
- O(10000) genes/genome
- O(1000) bases/gene
- Approximately 1% of DNA codes for genes (103104/109)
2
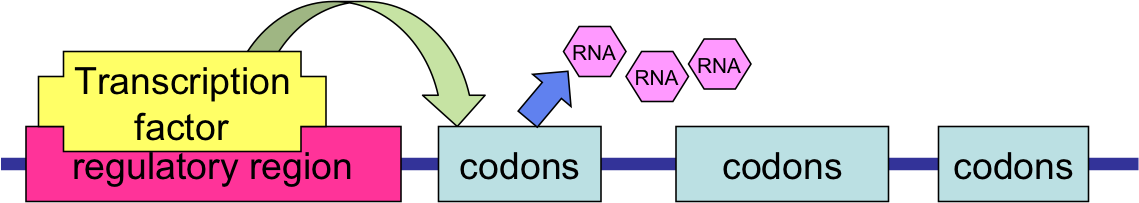
Regulatory Regions¶
- RNA polymerases seek out regulatory or promoting regions located 100-1000 bp upstream from the coding region
- They work in conjunction with special proteins called transcription factors (TFs) whose presence enables gene expression
- Within these regions are the Transcription Factor Binding Sites (TFBS), special DNA sequence patterns known as motifs that are specific to a given transcription factor
- A Single TF can influence the expression of many genes. Through biological experiments one can infer, at least a subset of these affected genes.
3
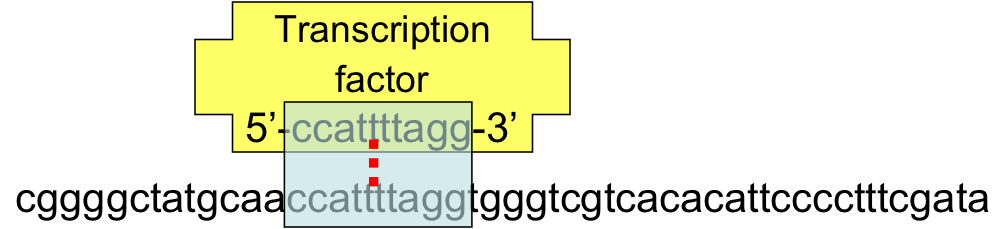
Transcription Factor Binding Sites¶
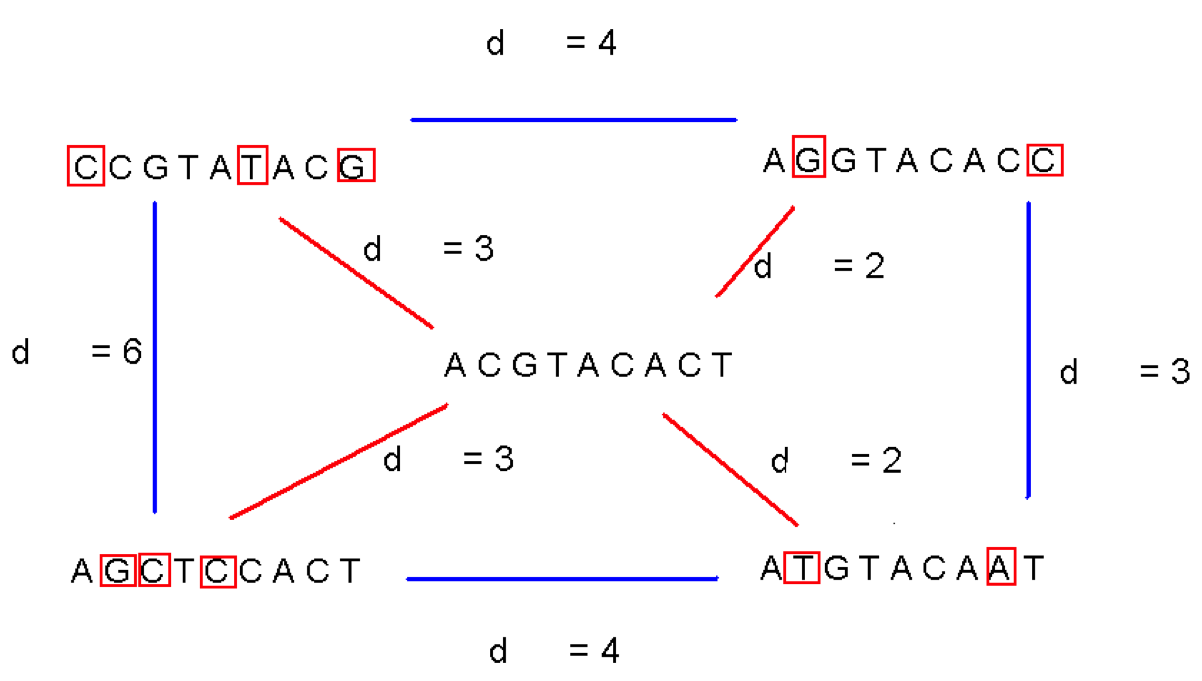
- A TFBS can be located anywhere within the regulatory region.
- TFBS may vary slightly across different regulatory regions since non-essential bases could mutate
- Transcription factors are robust (they will still bind) in the presence of small sequence differences by a few bases
4
Identifying Motifs: Complications¶
- We don’t know the motif sequence for every TF

- We don’t know where it is located relative to a gene’s start
- Moreover, motifs can differ slightly from one gene to the next
- We only know that it occurs somewhere near genes that share a TF
- How to discern a Motif’s frequent similar pattern from random patterns?
- How is this problem different that finding frequent k-mers from Lecture 2?
5
Let's look for an Easy Motif¶
1 tagtggtcttttgagtgtagatccgaagggaaagtatttccaccagttcggggtcacccagcagggcagggtgacttaat
2 cgcgactcggcgctcacagttatcgcacgtttagaccaaaacggagttagatccgaaactggagtttaatcggagtcctt
3 gttacttgtgagcctggttagatccgaaatataattgttggctgcatagcggagctgacatacgagtaggggaaatgcgt
4 aacatcaggctttgattaaacaatttaagcacgtagatccgaattgacctgatgacaatacggaacatgccggctccggg
5 accaccggataggctgcttattagatccgaaaggtagtatcgtaataatggctcagccatgtcaatgtgcggcattccac
6 tagatccgaatcgatcgtgtttctccctctgtgggttaacgaggggtccgaccttgctcgcatgtgccgaacttgtaccc
7 gaaatggttcggtgcgatatcaggccgttctcttaacttggcggtgtagatccgaacgtctctggaggggtcgtgcgcta
8 atgtatactagacattctaacgctcgcttattggcggagaccatttgctccactacaagaggctactgtgtagatccgaa
9 ttcttacacccttctttagatccgaacctgttggcgccatcttcttttcgagtccttgtacctccatttgctctgatgac
10 ctacctatgtaaaacaacatctactaacgtagtccggtctttcctgatctgccctaacctacaggtagatccgaaattcg
How would you go about finding a 10-mer that appears in every one of these strings?
6
Sneak Peek at the Answer¶
1 tagtggtcttttgagtgTAGATCCGAAgggaaagtatttccaccagttcggggtcacccagcagggcagggtgacttaat
2 cgcgactcggcgctcacagttatcgcacgtttagaccaaaacggagtTAGATCCGAAactggagtttaatcggagtcctt
3 gttacttgtgagcctggtTAGATCCGAAatataattgttggctgcatagcggagctgacatacgagtaggggaaatgcgt
4 aacatcaggctttgattaaacaatttaagcacgTAGATCCGAAttgacctgatgacaatacggaacatgccggctccggg
5 accaccggataggctgcttatTAGATCCGAAaggtagtatcgtaataatggctcagccatgtcaatgtgcggcattccac
6 TAGATCCGAAtcgatcgtgtttctccctctgtgggttaacgaggggtccgaccttgctcgcatgtgccgaacttgtaccc
7 gaaatggttcggtgcgatatcaggccgttctcttaacttggcggtgTAGATCCGAAcgtctctggaggggtcgtgcgcta
8 atgtatactagacattctaacgctcgcttattggcggagaccatttgctccactacaagaggctactgtgTAGATCCGAA
9 ttcttacacccttcttTAGATCCGAAcctgttggcgccatcttcttttcgagtccttgtacctccatttgctctgatgac
10 ctacctatgtaaaacaacatctactaacgtagtccggtctttcctgatctgccctaacctacaggTAGATCCGAAattcg
Now that you've seen the answer, how would you find it?
7
Let's meet Mr Brute Force¶

- He's often the best starting point when approaching a problem
- He'll also serve as a straw-man when designing new approaches
- Though he's seldom elegant, he gets the job done
- Often, we can't afford to wait for him
For our current problem a brute force solution would consider every k-mer position in all strings and see if they match. Given M sequences of length N, there are:
$$(N-k+1)^M$$position combinations to consider.
8
A Library of Helper Functions¶
- There's a tendancy to approach this problem with a series of nested for-loops, while the approach is valid, it doesn't generalize. It assumes a specific number of sequences.
- What we need is an iterator that generates all permutations of a sequence.
- This nested-for-loop iterator is called a Cartesian Product over sets.
- Python has a library to accomplish this:
import itertools
for number in itertools.product("01", repeat=3):
print ''.join(number)
for number in itertools.product(range(3), repeat=3):
print number,
for section in itertools.product(("I", "II", "III", "IV"),"ABC",range(1,3)):
print section
9
Now Let's Try Some Code¶
sequences = [
'tagtggtcttttgagtgtagatccgaagggaaagtatttccaccagttcggggtcacccagcagggcagggtgacttaat',
'cgcgactcggcgctcacagttatcgcacgtttagaccaaaacggagttagatccgaaactggagtttaatcggagtcctt',
'gttacttgtgagcctggttagatccgaaatataattgttggctgcatagcggagctgacatacgagtaggggaaatgcgt',
'aacatcaggctttgattaaacaatttaagcacgtagatccgaattgacctgatgacaatacggaacatgccggctccggg',
'accaccggataggctgcttattagatccgaaaggtagtatcgtaataatggctcagccatgtcaatgtgcggcattccac',
'tagatccgaatcgatcgtgtttctccctctgtgggttaacgaggggtccgaccttgctcgcatgtgccgaacttgtaccc',
'gaaatggttcggtgcgatatcaggccgttctcttaacttggcggtgtagatccgaacgtctctggaggggtcgtgcgcta',
'atgtatactagacattctaacgctcgcttattggcggagaccatttgctccactacaagaggctactgtgtagatccgaa',
'ttcttacacccttctttagatccgaacctgttggcgccatcttcttttcgagtccttgtacctccatttgctctgatgac',
'ctacctatgtaaaacaacatctactaacgtagtccggtctttcctgatctgccctaacctacaggtagatccgaaattcg']
def bruteForce(dna,k):
t = len(dna)
N = len(dna[0])
for offset in itertools.product(range(N-k+1), repeat=t):
for i in xrange(1,len(offset)):
if dna[0][offset[0]:offset[0]+k] != dna[i][offset[i]:offset[i]+k]:
break
else:
print offset, dna[0][offset[0]:offset[0]+10]
%time bruteForce(sequences[0:4], 10)
# you can try [0:5], but be prepared to wait
10
Approximate Matching¶
Now let's consider a more realistic motif finding problem, where the binding sites do not need to match exactly.
1 tagtggtcttttgagtgTAGATCTGAAgggaaagtatttccaccagttcggggtcacccagcagggcagggtgacttaat
2 cgcgactcggcgctcacagttatcgcacgtttagaccaaaacggagtTGGATCCGAAactggagtttaatcggagtcctt
3 gttacttgtgagcctggtTAGACCCGAAatataattgttggctgcatagcggagctgacatacgagtaggggaaatgcgt
4 aacatcaggctttgattaaacaatttaagcacgTAAATCCGAAttgacctgatgacaatacggaacatgccggctccggg
5 accaccggataggctgcttatTAGGTCCAAAaggtagtatcgtaataatggctcagccatgtcaatgtgcggcattccac
6 TAGATTCGAAtcgatcgtgtttctccctctgtgggttaacgaggggtccgaccttgctcgcatgtgccgaacttgtaccc
7 gaaatggttcggtgcgatatcaggccgttctcttaacttggcggtgCAGATCCGAAcgtctctggaggggtcgtgcgcta
8 atgtatactagacattctaacgctcgcttattggcggagaccatttgctccactacaagaggctactgtgTAGATCCGTA
9 ttcttacacccttcttTAGATCCAAAcctgttggcgccatcttcttttcgagtccttgtacctccatttgctctgatgac
10 ctacctatgtaaaacaacatctactaacgtagtccggtctttcctgatctgccctaacctacaggTCGATCCGAAattcg
Actually, none of the sequences have an unmodified copy of the original motif
11
Profile and Consensus¶
- Align candidate motifs by their start indexes $$s = (s_1, s_2, …, s_t)$$
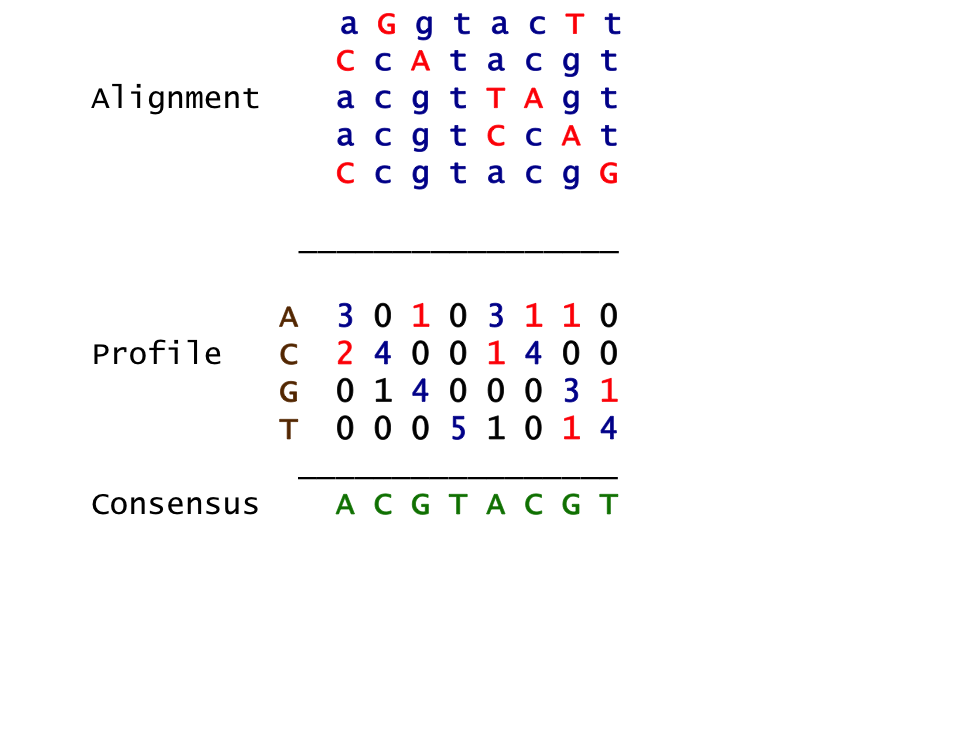
- Construct a matrix profile with the frequencies of each nucleotide in columns
- Consensus nucleotide in each position has the highest score in column
12
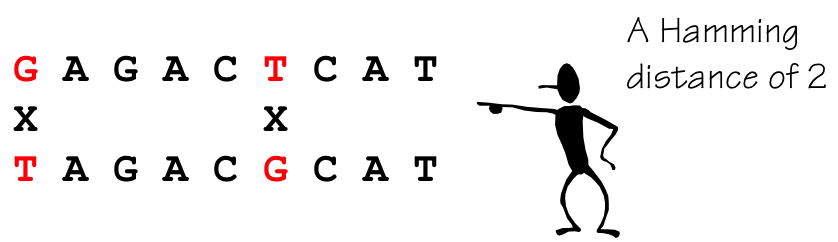
Consensus¶
- One can think of the consensus as an ancestor motif, from which mutated motifs emerged
- The distance between an actual motif and the consensus sequence is generally less than that for any two actual motifs
- Hamming distance is number of positions that differ between two strings
13
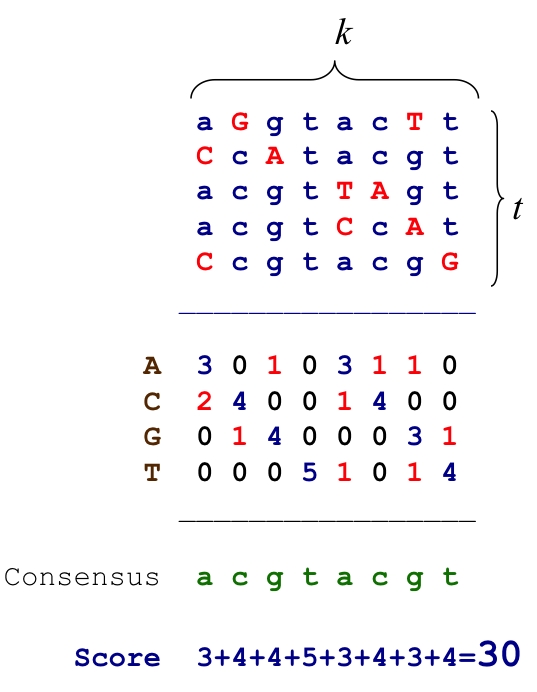
Scoring Motifs¶
- Given $s = (s_1, s_2, …, s_t)$ and DNA $$Score(s,DNA) = \sum_{i=1}^{k} \max_{j \in {A,C,G,T}} count(j,i)$$
- So our approach is back to brute force
- We consider every candidate motif in every string
- Return the set of indices with the highest score
15
Let's try again, and handle errors this time!¶
seqApprox = [
'tagtggtcttttgagtgtagatctgaagggaaagtatttccaccagttcggggtcacccagcagggcagggtgacttaat',
'cgcgactcggcgctcacagttatcgcacgtttagaccaaaacggagttggatccgaaactggagtttaatcggagtcctt',
'gttacttgtgagcctggttagacccgaaatataattgttggctgcatagcggagctgacatacgagtaggggaaatgcgt',
'aacatcaggctttgattaaacaatttaagcacgtaaatccgaattgacctgatgacaatacggaacatgccggctccggg',
'accaccggataggctgcttattaggtccaaaaggtagtatcgtaataatggctcagccatgtcaatgtgcggcattccac',
'tagattcgaatcgatcgtgtttctccctctgtgggttaacgaggggtccgaccttgctcgcatgtgccgaacttgtaccc',
'gaaatggttcggtgcgatatcaggccgttctcttaacttggcggtgcagatccgaacgtctctggaggggtcgtgcgcta',
'atgtatactagacattctaacgctcgcttattggcggagaccatttgctccactacaagaggctactgtgtagatccgta',
'ttcttacacccttctttagatccaaacctgttggcgccatcttcttttcgagtccttgtacctccatttgctctgatgac',
'ctacctatgtaaaacaacatctactaacgtagtccggtctttcctgatctgccctaacctacaggtcgatccgaaattcg']
def Score(s, DNA, k):
"""
compute the consensus SCORE of a given k-mer
alignment given offsets into each DNA string.
s = list of starting indices
DNA = list of nucleotide strings
k = Target Motif length
"""
score = 0
for i in xrange(k):
# loop over string positions
cnt = dict(zip("acgt",(0,0,0,0)))
for j, sval in enumerate(s):
base = DNA[j][sval+i]
cnt[base] += 1
score += max(cnt.values())
return score
def BruteForceMotifSearch(dna,k):
t = len(dna)
N = len(dna[0])
bestScore = 0
bestAlignment = []
for offset in itertools.product(range(N-k+1), repeat=t):
s = Score(offset,dna,k)
if (s > bestScore):
bestAlignment = [p for p in offset]
bestScore = s
print bestAlignment, bestScore
%timeit Score([17, 47, 18, 33, 21, 0, 46, 70, 16, 65], seqApprox, 10)
%time BruteForceMotifSearch(seqApprox[0:4], 10)
16
Running Time of BruteForceMotifSearch¶
- Search (N - k + 1) positions in each of t sequences, by examining (N - k + 1)t sets of starting positions
- For each set of starting positions, the scoring function makes O(tk) operations, so complexity is
$$t k (N – k + 1)^t = O(t k N^t)$$

- That means that for t = 10, N = 80, k = 10 we must perform approximately $10^{21}$ computations
- Generously assuming $10^9$ comps/sec it will require only $10^{12}$ secs $\frac{10^{12}}{(60 * 60 * 24 *365)} > 30000 $ years
- Want to wait?
17
How conservative is this estimate?¶
- For the example we just did t = 4, N = 80, k = 10

- So that gives ≈ $4.0 \times 10^9$ operations
- Using our $10^9$ operations per second estimate, it should have taken only 4 secs.
- Instead it took closer to 700 secs, which suggests we are getting around 5.85 million operations per second.
- So, in reality it will even take longer!
18
How can we find Motifs in our lifetime?¶
- Should we give up on Python and write in C? Assembly Language?
- Will biological insights save us this time?
- Are there other ways to find Motifs?
- Consider that if you knew what motif you were looking for, it would take only $$t k (N – k + 1) t = O(t^2 k N)$$
- Is that significantly better?
19