| 2_4 Female Full X Chromosome Results | shunping / Version 20 |
The following statistics are computed using only female FG, FF, and GG samples.
| 606191 | Number of high confidence sanger SNPs on X informative between F (CAST) and G (PWK) | |
| 24381 24208 | Number of X SNP positions with pileup heights > 50 reads in all 6 FG F1s | |
| 21175 21066 | Number of X SNP positions with pileup heights > 50 reads in all 6 F inbred samples (Note: there is one bad F sample) |
|
| 30454 30266 | Number of X SNP positions with pileup heights > 50 reads in all 6 G inbred samples |
|
| 11342 11266 | Number of X SNP positions with consistent genotypes (> 75% in 4 or more samples) in F inbred samples | |
| 16265 16127 | Number of X SNP positions with consistent genotypes (> 75% in 4 or more samples) in G inbred samples | |
| 1706 1643 | Intersection of SNP positions common to FG, F, and G with > 50 reads and having consistent F and G genotypes within coresponding inbreds |
Allelic ratio at informative SNP positions with pileups > 50 (24381 24208)
Allelic ratio at subset of SNP positions with consistent inbred genotypes (1706 1643)
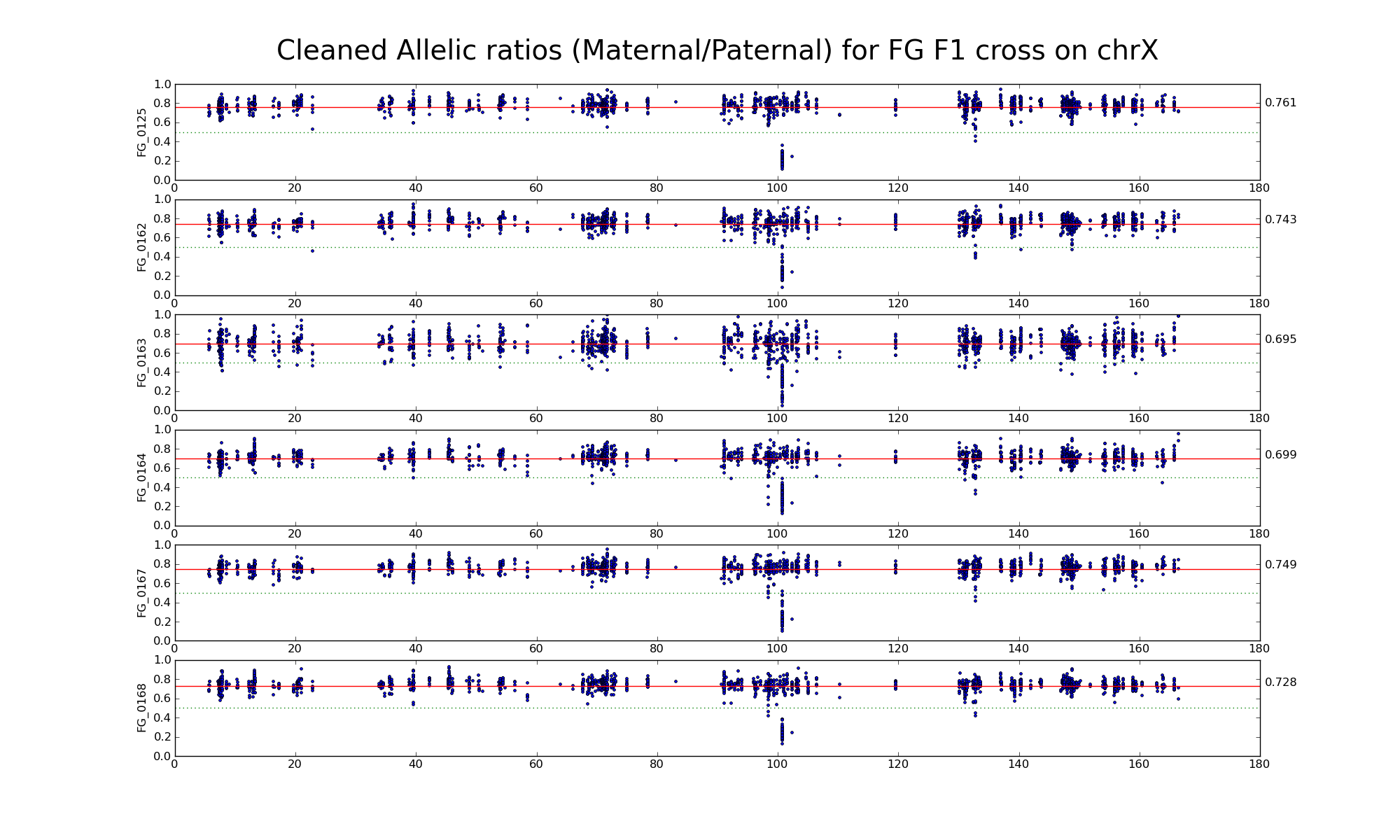
Is there an X-inactivation escapee around 135 Mb?
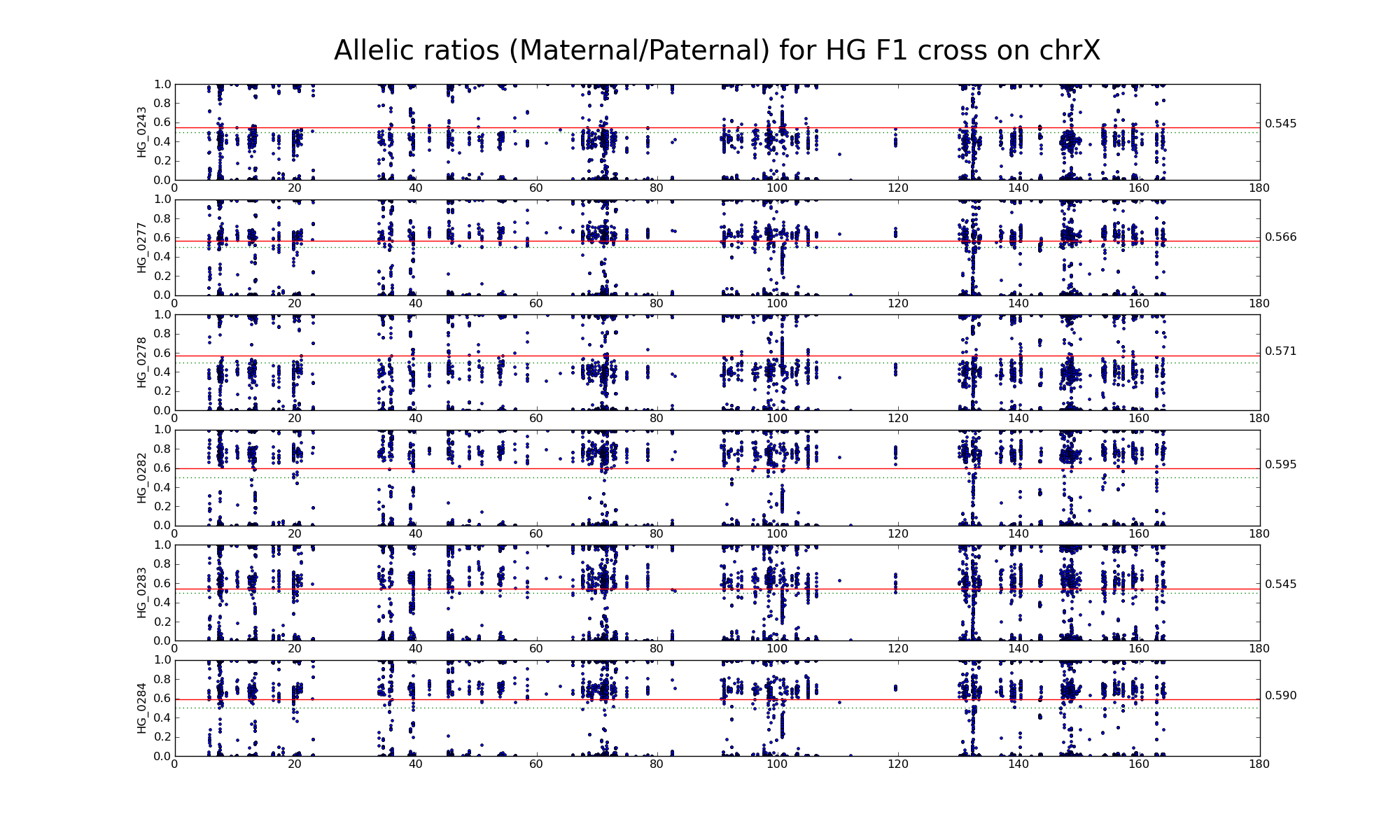
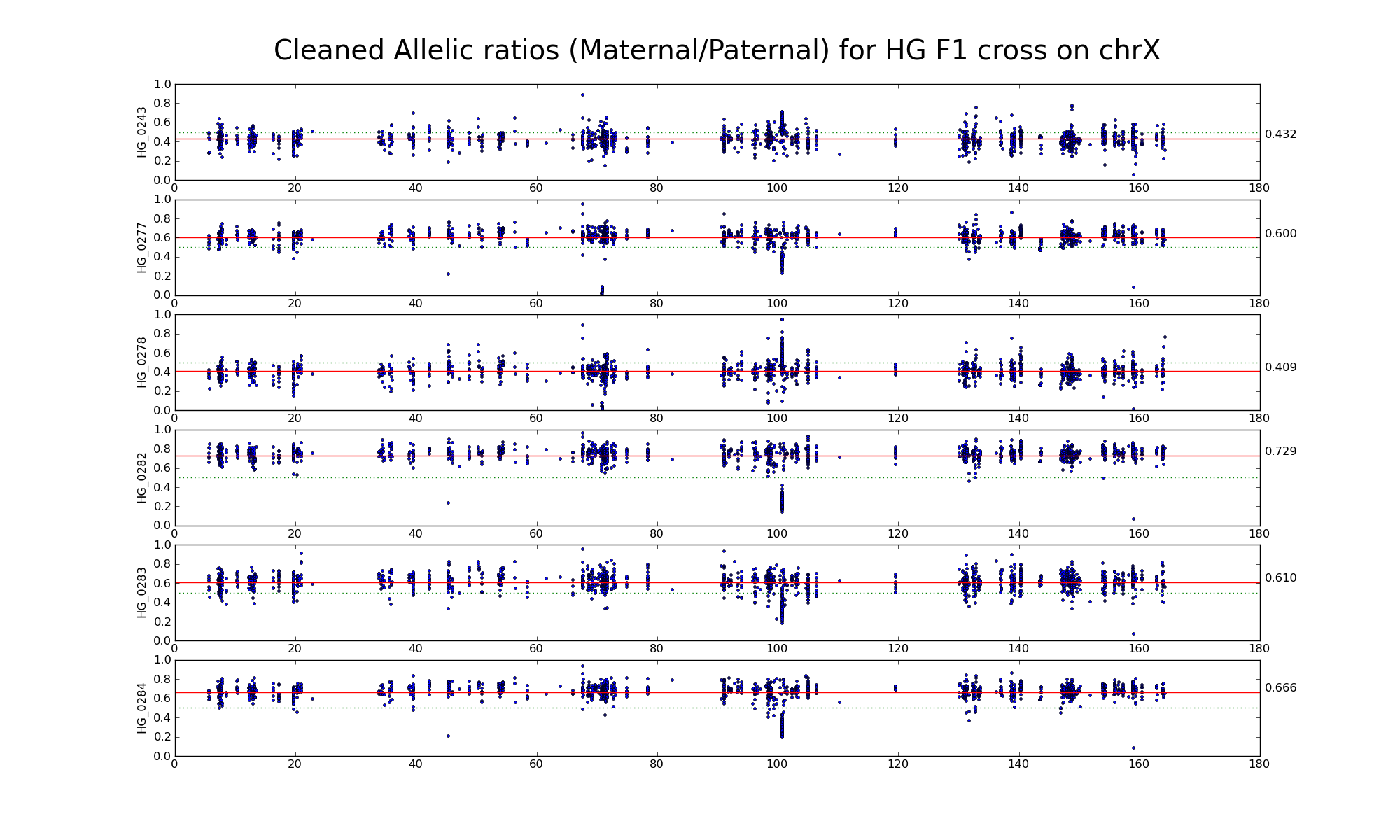
The following statistics are computed using only female GF, FF, and GG samples.
| 606191 | Number of high confidence sanger SNPs on X informative between G (PWK) and F (CAST) | |
| 30452 30265 | Number of X SNP positions with pileup heights > 50 reads in all 6 GF F1s | |
| 30454 30266 | Number of X SNP positions with pileup heights > 50 reads in all 6 G inbred samples |
|
| 21175 21066 | Number of X SNP positions with pileup heights > 50 reads in all 6 F inbred samples (Note: there is one bad F sample) |
|
| 15592 15458 | Number of X SNP positions with consistent genotypes (> 75% in 4 or more samples) in G inbred samples | |
| 11786 11710 | Number of X SNP positions with consistent genotypes (> 75% in 4 or more samples) in F inbred samples | |
| 1720 1668 | Intersection of SNP positions common to GF, G, and F with > 50 reads and having consistent G and F genotypes within coresponding inbreds |
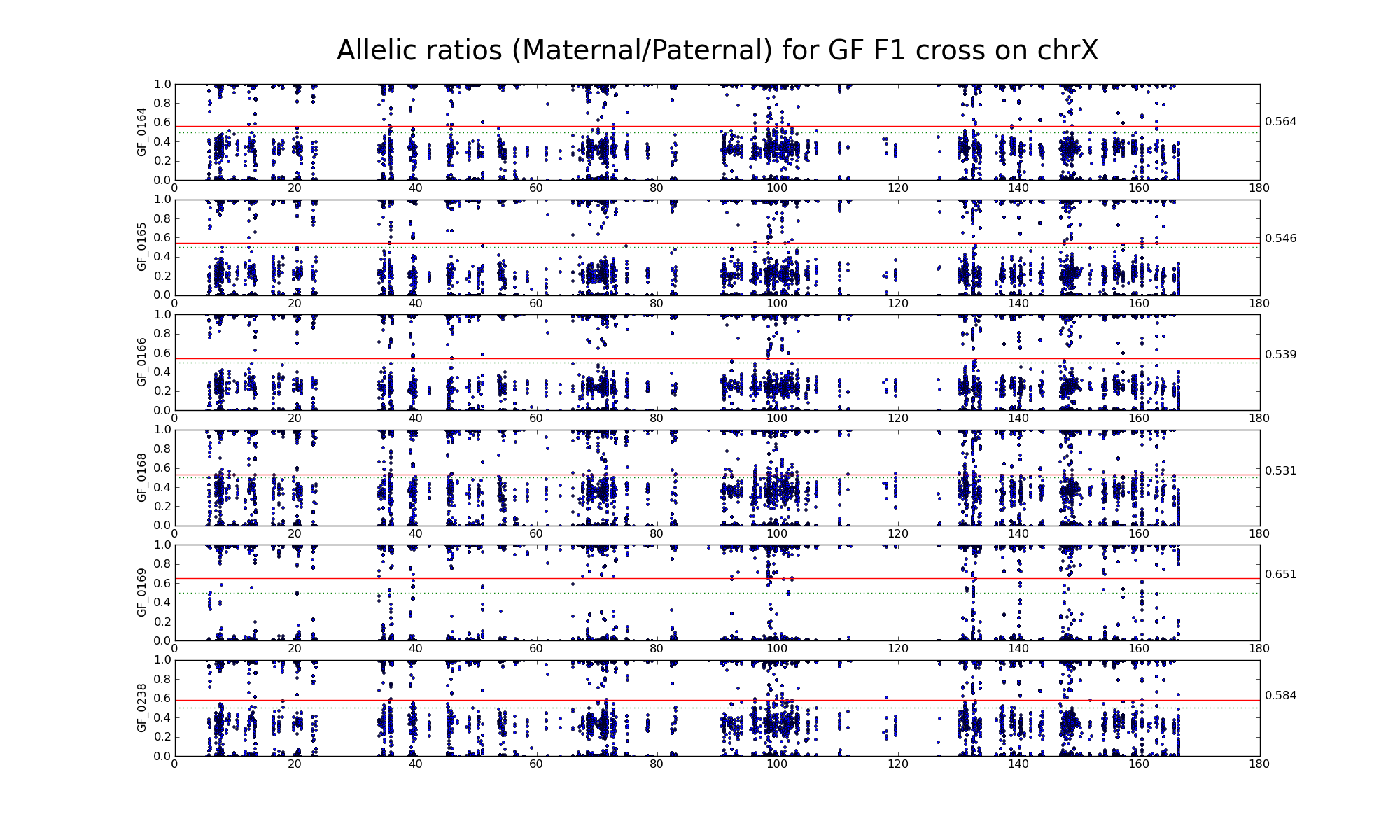
Allelic ratio at informative SNP positions with pileups > 50 (30452 30265)
Allelic ratio at subset of SNP positions with consistent inbred genotypes (1720 1668)
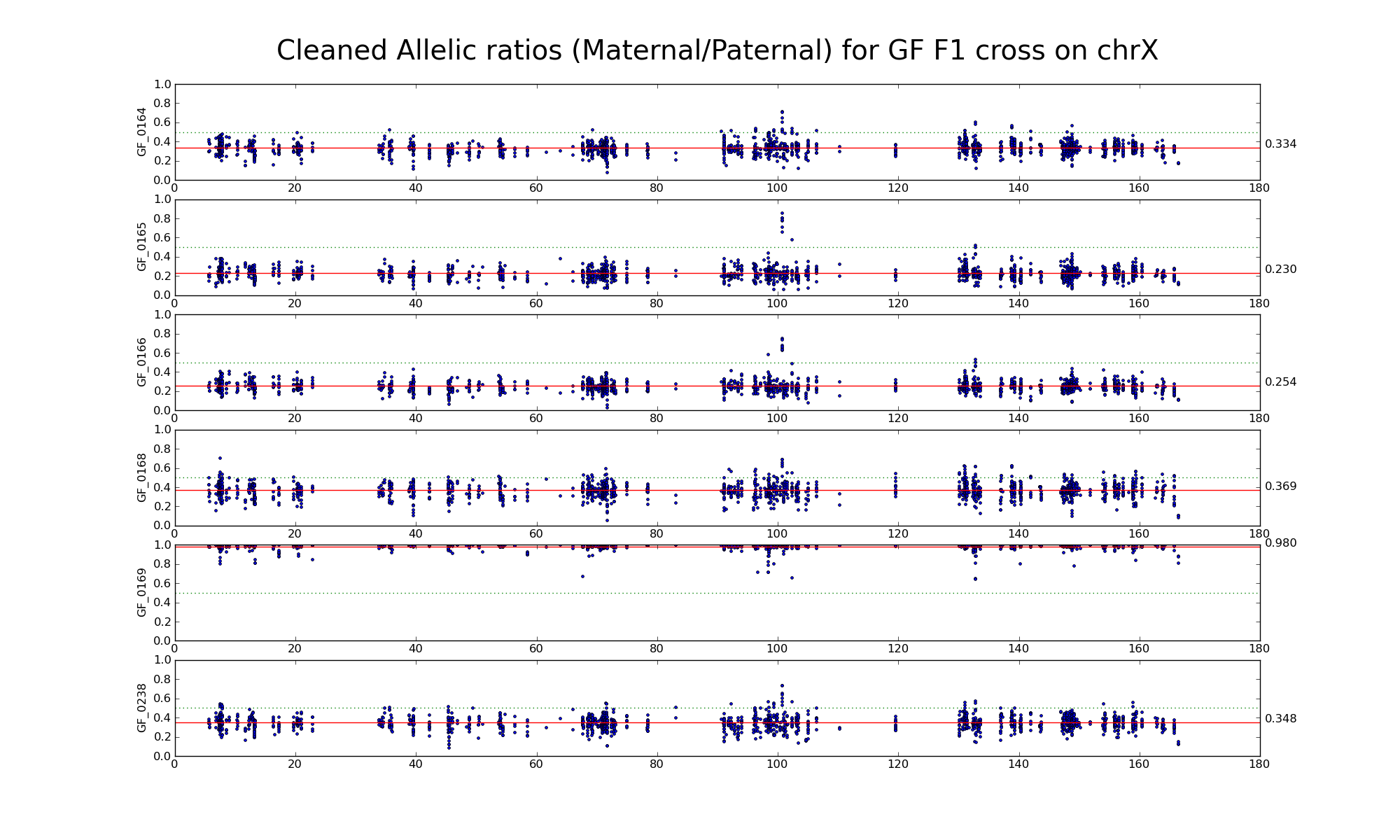
Note: Sample GF_0169 appears "male-like".
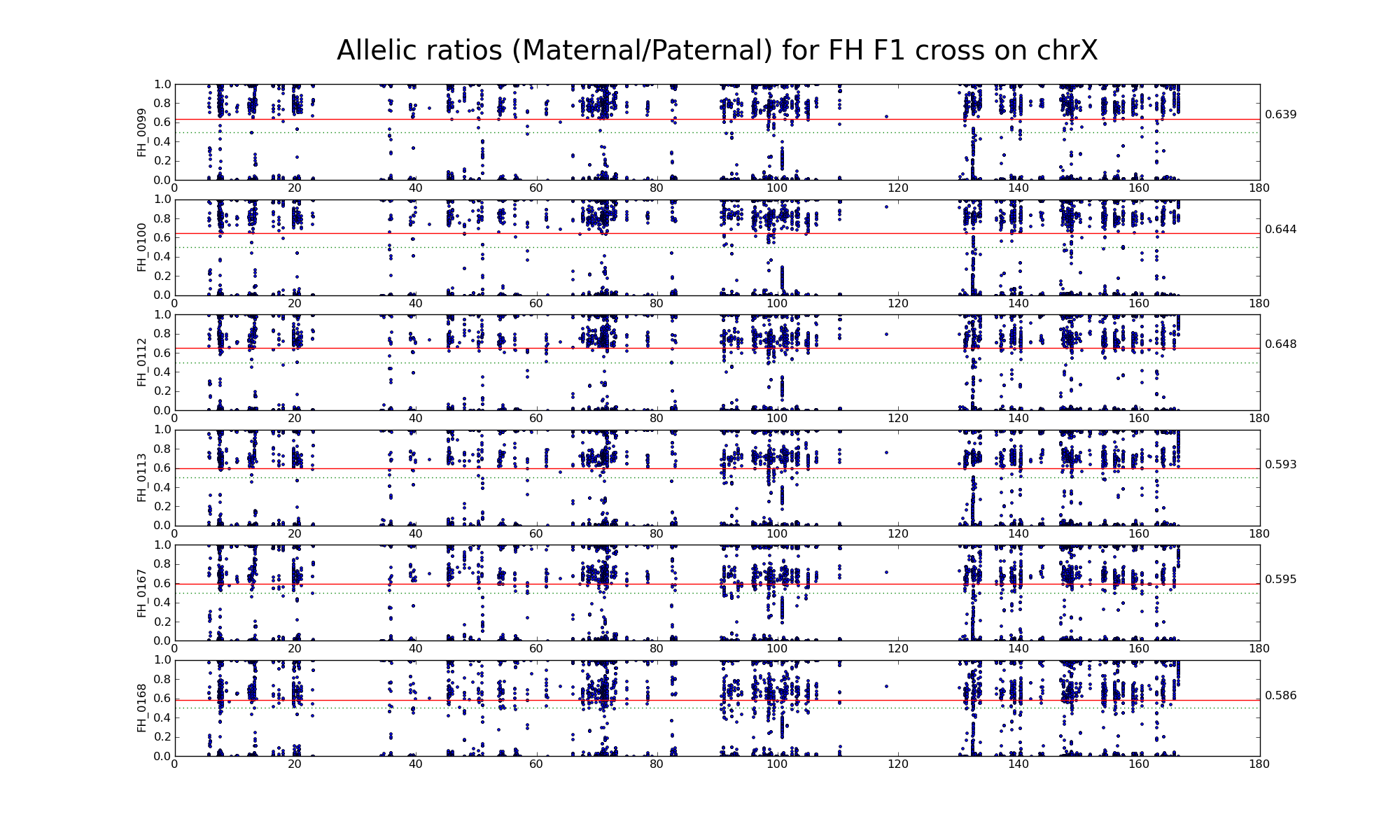
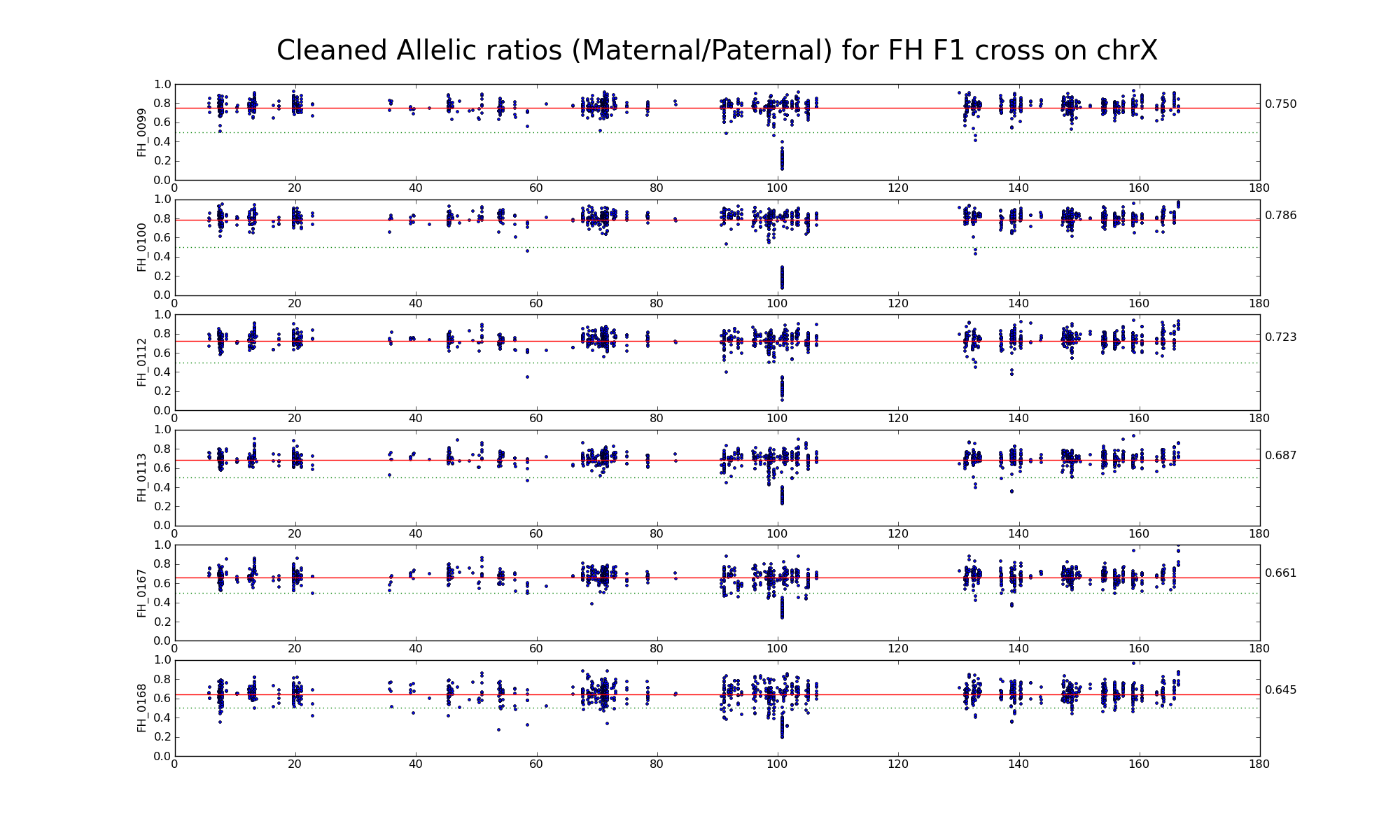
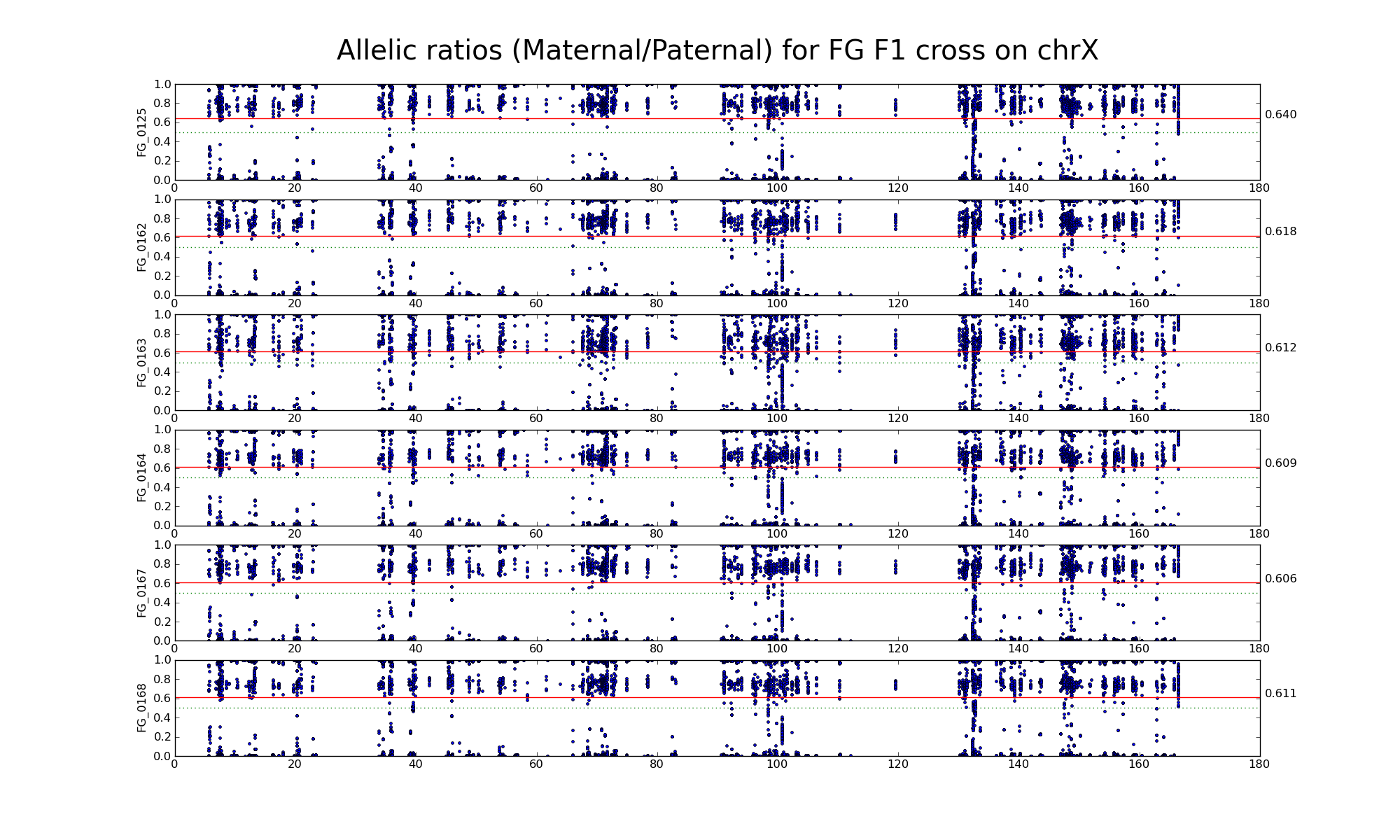
The following statistics are computed using only female FH, FF, and HH samples.
| 502333 | Number of high confidence sanger SNPs on X informative between F (CAST) and H (WSB) | |
| 20750 20660 | Number of X SNP positions with pileup heights > 50 reads in all 6 FH F1s | |
| 19793 19719 | Number of X SNP positions with pileup heights > 50 reads in all 6 F inbred samples (Note: there is one bad F sample) |
|
| 26762 26650 | Number of X SNP positions with pileup heights > 50 reads in all 6 H inbred samples |
|
| 10577 10523 | Number of X SNP positions with consistent genotypes (> 75% in 4 or more samples) in F inbred samples | |
| 14182 14103 | Number of X SNP positions with consistent genotypes (> 75% in 4 or more samples) in H inbred samples | |
| 1474 1444 | Intersection of SNP positions common to FH, F, and H with > 50 reads and having consistent F and H genotypes within coresponding inbreds |
Allelic ratio at informative SNP positions with pileups > 50 (20750 20660)
Allelic ratio at subset of SNP positions with consistent inbred genotypes (1474 1444)
The following statistics are computed using only female HF, HH, and FF samples.
| 502333 | Number of high confidence sanger SNPs on X informative between H (WSB) and F (CAST) | |
| 23040 22931 | Number of X SNP positions with pileup heights > 50 reads in all 6 HF F1s | |
| 26762 26650 | Number of X SNP positions with pileup heights > 50 reads in all 6 H inbred samples |
|
| 19793 19719 |
Number of X SNP positions with pileup heights > 50 reads in all 6 F inbred samples (Note: there is one bad F sample) |
|
| 13602 13523 | Number of X SNP positions with consistent genotypes (> 75% in 4 or more samples) in H inbred samples | |
| 11034 10980 | Number of X SNP positions with consistent genotypes (> 75% in 4 or more samples) in F inbred samples | |
| 1532 1502 | Intersection of SNP positions common to HF, H, and F with > 50 reads and having consistent H and F genotypes within coresponding inbreds |
Allelic ratio at informative SNP positions with pileups > 50 (23040 22931)
Allelic ratio at subset of SNP positions with consistent inbred genotypes (1532 1502)
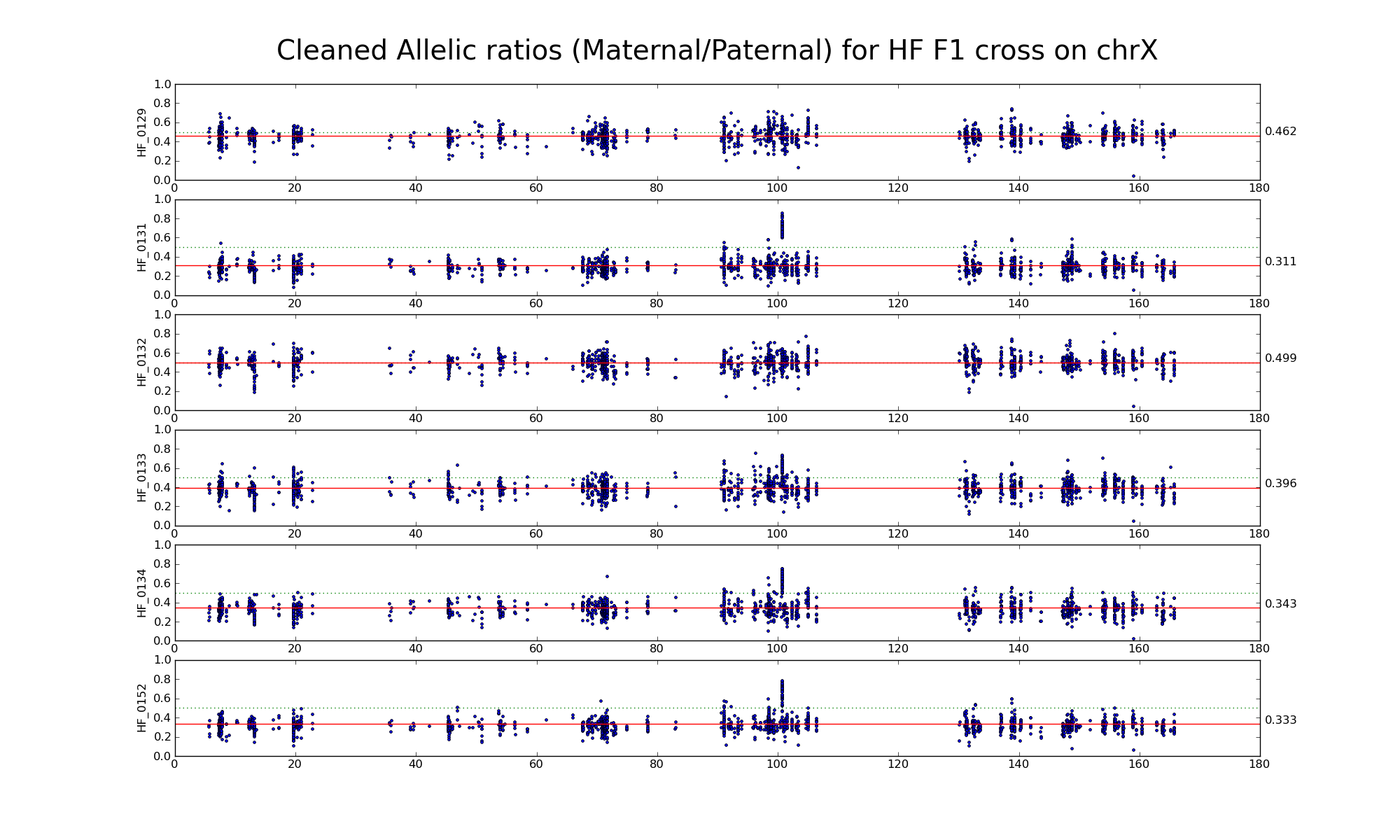
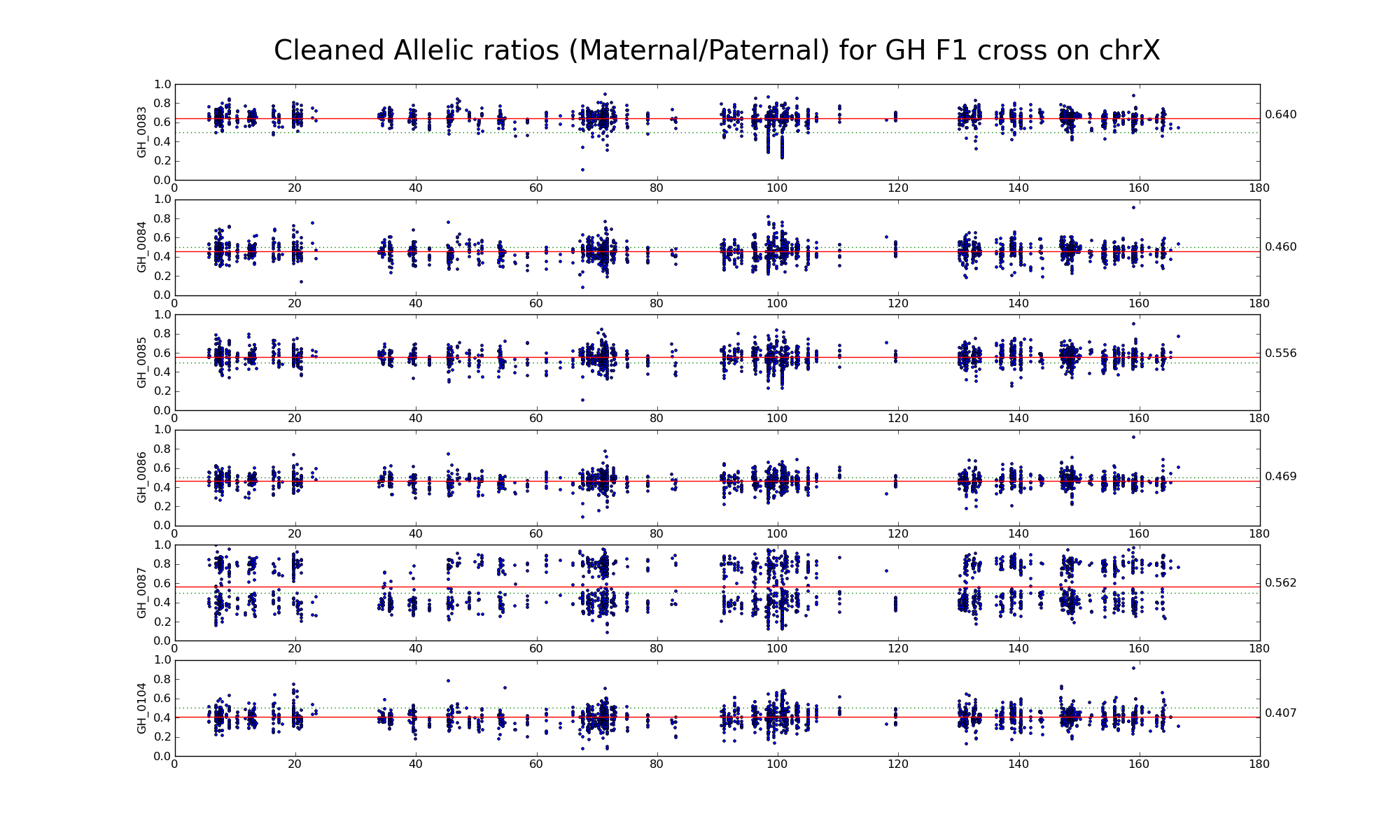
The following statistics are computed using only female GH, GG, and HH samples.
| 656999 | Number of high confidence sanger SNPs on X informative between G (PWK) and H (WSB) | |
| 36941 36688 | Number of X SNP positions with pileup heights > 50 reads in all 6 GH F1s | |
| 33169 32923 | Number of X SNP positions with pileup heights > 50 reads in all 6 G inbred samples |
|
| 31054 30828 | Number of X SNP positions with pileup heights > 50 reads in all 6 H inbred samples |
|
| 17370 17207 | Number of X SNP positions with consistent genotypes (> 75% in 4 or more samples) in G inbred samples | |
| 16912 16751 | Number of X SNP positions with consistent genotypes (> 75% in 4 or more samples) in H inbred samples | |
| 2317 2230 | Intersection of SNP positions common to GH, G, and H with > 50 reads and having consistent G and H genotypes within coresponding inbreds |
Allelic ratio at informative SNP positions with pileups > 50 (36941 36688)
Allelic ratio at subset of SNP positions with consistent inbred genotypes (2317 2230)
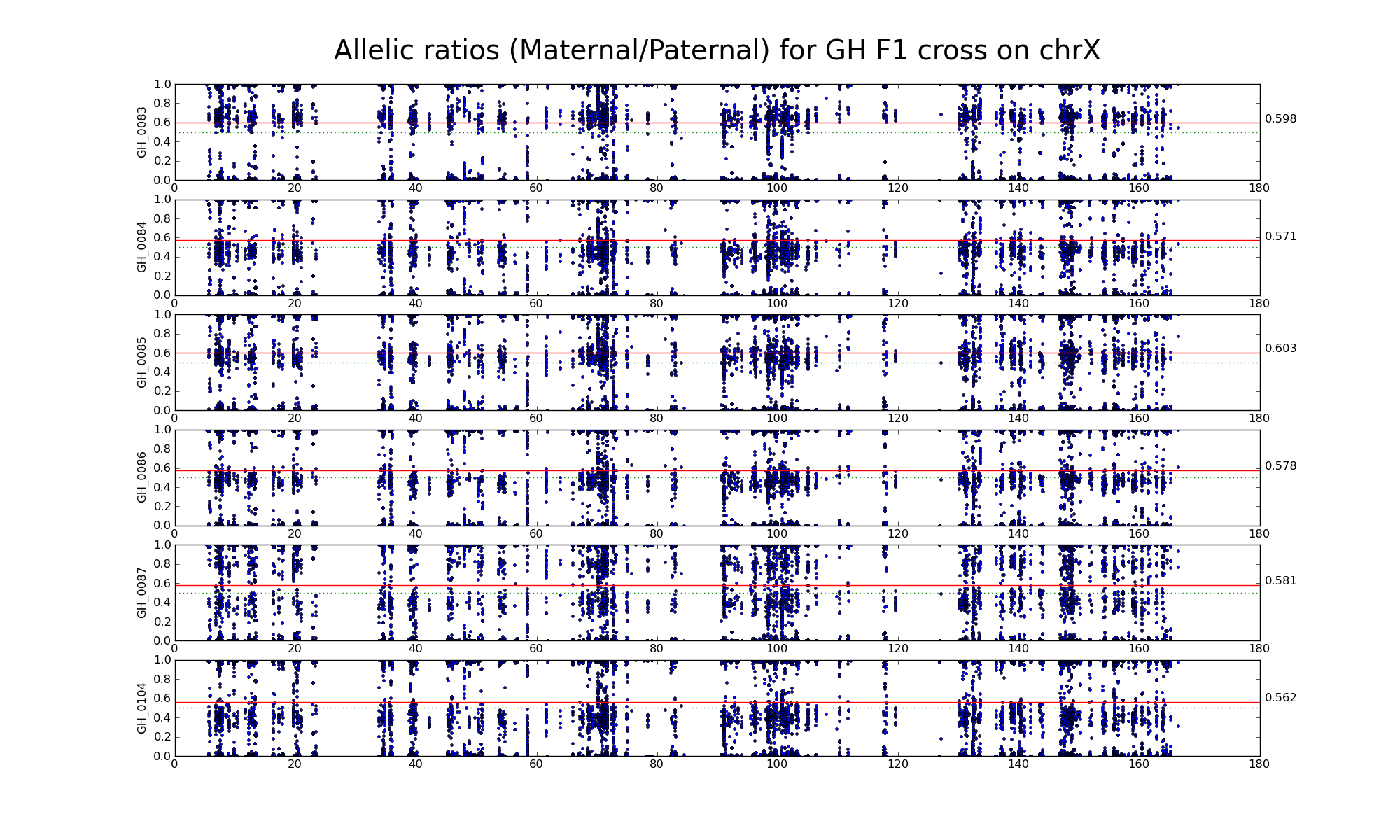
The following statistics are computed using only female HG, HH, and GG samples.
| 656999 | Number of high confidence sanger SNPs on X informative between H (WSB) and G (PWK) | |
| 18581 18460 | Number of X SNP positions with pileup heights > 50 reads in all 6 HG F1s | |
| 31054 30828 | Number of X SNP positions with pileup heights > 50 reads in all 6 H inbred samples |
|
| 33169 32923 |
Number of X SNP positions with pileup heights > 50 reads in all 6 G inbred samples |
|
| 16307 16146 | Number of X SNP positions with consistent genotypes (> 75% in 4 or more samples) in H inbred samples | |
| 18033 17866 | Number of X SNP positions with consistent genotypes (> 75% in 4 or more samples) in G inbred samples | |
| 1689 1636 | Intersection of SNP positions common to HG, H, and G with > 50 reads and having consistent H and G genotypes within coresponding inbreds |
Allelic ratio at informative SNP positions with pileups > 50 (18581 18460)
Allelic ratio at subset of SNP positions with consistent inbred genotypes (1689 1636)